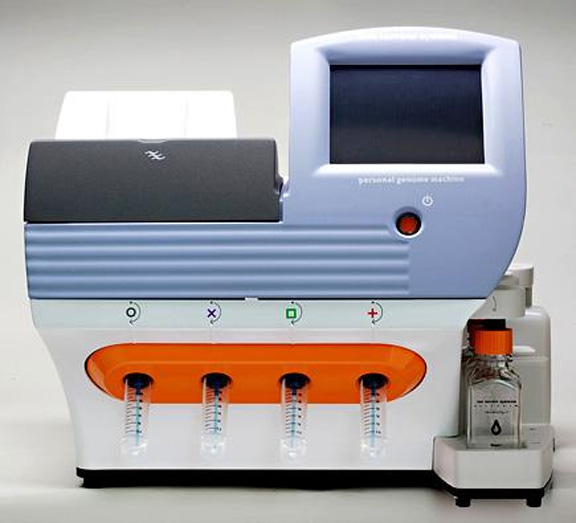
NEW YORK, (Reuters) – After years of predictions that the “$1,000 genome” – a read-out of a person’s complete genetic information for about the cost of a dental crown – was just around the corner, a U.S. company is announcing that it has achieved that milestone and taken the technology several steps ahead.
 The new genome-sequencing machine from Ion Torrent, a division of Life Technologies Corp., in Guilford, Connecticut, is 1,000 times more powerful than existing technology, says CEO and chairman Jonathan Rothberg.
The new genome-sequencing machine from Ion Torrent, a division of Life Technologies Corp., in Guilford, Connecticut, is 1,000 times more powerful than existing technology, says CEO and chairman Jonathan Rothberg.
Taking up about as much space as an office printer, it can sequence an entire genome in a single day rather than six to eight weeks required only a few years ago. The new sequencer, says cardiologist Eric Topol, chief academic officer of private California hospital and doctor network Scripps Health, “represents an exceptional advance and can change medicine.”
Ion Torrent will sell the tabletop machine, called the Ion Proton Sequencer, for $99,000 to $149,000, making it affordable for large medical practices or clinics; existing sequencers cost up to $750,000. The computer chip and biochemicals to sequence a genome will cost $1,000, compared to, for example, $3,000 to test for mutations just in the BRCA genes that raise the risk of breast and ovarian cancer and $5,000 for a complete genome sequencing by Ion Torrent competitor Illumina Inc.
For a graphic on the shrinking cost of genome sequencing, see: http://link.reuters.com/xys85s
For now, Rothberg expects research labs to be his main customers, using Proton to obtain the complete genome sequence of people with cancer or autism, for instance, and thereby elucidate a disease’s underlying genetic causes as well as possible ways to treat it. The company has signed on Baylor College of Medicine, Yale School of Medicine and the Broad Institute as its first customers.
Other scientists and physicians, however, say the long-awaited arrival of the $1,000 genome opens the door to widespread whole-genome sequencing even of people who are not ill. And that raises ethical, legal, and medical issues that experts are only beginning to grapple with.
“I’m a big proponent of bringing genetics into the clinic,” says Thomas Quertermous, chief of the division of cardiovascular medicine at Stanford University and an expert in the genetics of heart disease. “But it has to be done in a timely way, and not before its time.”
Babies might be first in line for whole-genome sequencing. Every state requires newborns to be screened for at least 29 genetic diseases.
“If the cost of whole-genome sequencing gets sufficiently low, you could sequence all the genes in a newborn” for less than the individual tests and follow-ups required when one comes back positive, says Richard Lifton, chairman of the genetics department at Yale University. “I’m increasingly confident that’s going to happen. But we need to be careful how we utilize this information. Do you tell a newborn’s parents his apoE status” — that is, whether he has the form of a gene that raises the risk of Alzheimer’s disease?
The cost of whole-genome sequencing will continue to plummet. Lifton foresees a “zero-dollar genome,” making it likely that “we will just do it as part of routine clinical care” for children and adults. A Yale team led by Murat Gunel has already used partial genome sequencing of the 1.5 percent of the genome, called the exome, that codes for proteins to determine the cause of a mysterious and still unnamed genetic disease that results in severe brain malformations.
Because no genes had been identified as causing the malformation, it was not possible to do a standard genetic test, which reveals whether a particular gene is normal or mutated. But exome sequencing showed that a previously unknown gene on chromosome 19 is responsible, he and colleagues reported in 2010. “The new Proton instrument is a big step up,” says Lifton. “It promises to markedly increase the speed and reduce the cost of genome-level sequencing.”
TSUNAMI OF DATA
The discovery of the mutation behind the mysterious genetic disorder demonstrated the advantage of whole-genome sequencing compared to single-gene tests, as scientists can’t test for a gene they don’t know exists. Beyond such uses, say experts, whole-genome sequencing might not be the medical miracle that proponents forecast.
One problem is that the costs only start with the actual sequencing. “The cost of understanding the sequence will be much, much higher,” says bioethicist Hank Greely of Stanford University. He participated in a 2010 project that sequenced the full genome of Stanford bioengineer Stephen Quake. The sequencing cost $48,000.
But because it found 2.6 million DNA misspellings and 752 other genetic glitches, says Greely, “it took a few hundred thousand dollars worth of labor from Ph.D. students and faculty working 4,000 to 5,000 hours to understand what the sequence meant” — that Quake had a higher-than-average risk of sudden cardiac death, a lower risk of Alzheimer’s, and a higher risk of prostate cancer.
Another challenge is that whole-genome sequencing generates a tsunami of data. It would take a genetic counselor some five hours to explain what a typical genome means, further adding to the true cost. The United States has about 2,500 genetic counselors, not nearly enough to meet the need if whole-genome sequencing becomes widespread. Might doctors take up the slack? “Surveys show that 90 percent of patients trust their physician to explain genomics data to them,” says Scripps’ Topol. “And 90 percent of physicians say they don’t feel comfortable with genomics data.”
Although many bioethicists focus on the psychological harm patients might suffer when DNA tests show an elevated risk of cancer, diabetes, Parkinson’s, and other diseases, genomics information could also threaten patients’ physical health if it is misconstrued. A woman whose DNA sequencing shows she does not carry BRCA mutations that raise her risk of breast cancer “might say, great, I don’t need mammograms,” says Stanford’s Greely. “But a negative BRCA test reduces her risk of breast cancer from 12 percent to 11.96 percent. My dread is less that patients will be damaged psychologically and more that they will misunderstand (genome sequence data) and do stupid things.”
Unlike tests that detect glitches in genes that a patient or physician asks to have checked (those that raise the risk of, say, colon cancer if that disease runs in the family), and unlike the dozens of genes that “personal genetics” companies test for, whole-genome sequencing reveals every bit of information the genome contains about diseases or traits.
Given the ubiquity of mutations, everyone carries genes that predispose them to more than one serious or lethal disease. Bioethicists are only beginning to study how that knowledge might affect someone’s decisions, from marrying or having children to saving for retirement.
Another challenge is that although a person’s genome doesn’t change, its meaning will. As scientists uncover more DNA variants that protect against disease and variants that make it more likely, a genome sequence that meant one thing in 2012 will have a different meaning in 2013, not to mention 2020.
A DNA variant that was once thought to be dangerous “might turn out to be benign if countered by another,” says Greely. “Whose responsibility will it be to tell you that, years later?” Today’s DNA testing companies offer subscriptions that give customers regular updates like that.
Geneticists are also still struggling with the fact that most of the risk genes raise the likelihood that the person will develop the disease only slightly. “The bottom line is, the effect size is so small it’s virtually insignificant clinically,” says Quertermous. “So how should doctors incorporate that knowledge into their armamentarium? They won’t be able to look at 6 billion data bits” – the amount in a whole-genome scan – “and evaluate what it means for patients.”
Knowing a patient’s whole-genome sequence, even if it raises the risk of diseases by only a few percent, might lead malpractice-wary doctors to order follow-up tests. If someone’s genome suggests an elevated risk of heart disease, for instance, a physician might feel compelled to order regular cardiac CT angiograms, which cost $1,500 or more.
That would not only raise health-care costs, but might put patients through a physically and psychologically onerous ordeal unnecessarily. “There is no evidence that ‘positive’ (DNA) tests, based only on the screening for common genetic variations, will justify a specific medical follow-up and procure a medical benefit to individuals,” argues geneticist Thierry Frebourg of University Hospital in Rouen, France in a commentary in an upcoming issue of the European Journal of Human Genetics. Instead, whole-genome sequencing might join the ranks of diagnostics, such as PSA tests for prostate cancer, that cost tens of millions of dollars a year but do not benefit patients, let alone save lives.
INEFFECTIVE DRUGS
Full-genome sequencing could provide real benefits in determining which patients will benefit from a drug. For instance, only half the hepatitis C patients who take Pegasys, a $50,000-a-year drug from Roche Holding AG’s Genentech, and half the rheumatoid arthritis patients who take $26,000-a-year Enbrel from Amgen Inc and Pfizer Inc , benefit from them, notes Scripps’ Topol, who analyzes the potential benefits of genomic medicine in his upcoming book, The Creative Destruction of Medicine.
Using genomic data to identify which patients will and will not benefit could save patients and insurers tens of billions of dollars a year now spent on ineffective drugs.
If genetic information causes patients to take better care of themselves – eating more healthfully if they carry genes that raise the risk of diabetes or heart disease, for instance – they can improve health. One 2010 study found that of people who bought direct-to-consumer genetic testing by companies such as 23andme, 34 percent said the results made them more careful about their diet and 14 percent exercised more.
Others incorrectly see DNA as destiny, and interpret an increased genetic risk of, say, obesity as a license to overeat, thinking they are fated to be fat. “Good” genes might lead to equally dangerous behavior. “A patient with hypertension might be told by his doctor, ‘I’ve looked at your DNA and you’re clean!’,” says Stanford’s Quertermous. “He might think, great, I don’t need to check my blood pressure anymore or even take my medication.”
As the science advances, however, the value of whole-genome sequencing to patients will grow. The common DNA variants that have been identified “account for only a small part of the heritability of disease,” says Kari Stefansson, founder, chairman, and CEO of deCode Genetics of Reykjavik, Iceland.
“The expectation is that a significant part of the missing heritability lies in rare variants, and to find those you have to do whole-genome sequencing.” deCode is sequencing the complete genomes of 3,750 Icelanders, and has so far identified rare variants with large effects on the risk of ovarian cancer, glioma, gout, and heart conditions that require a pacemaker. Those discoveries would have been difficult or impossible without whole-genome sequencing.
Whole-genome sequencing also promises to address one of the most troubling problems with current DNA tests, which probe some of the 1,500 or so genes that have been associated with disease out of a total of 22,000 human genes. But scientists do not know how disease risk is raised or lowered by “moderator genes,” which affect other genes. “Do we know how combinations of genes affect risk?” Stanford’s Quertermous asks. “The answer is completely no.” As a result, the disease risk that is calculated from current genetic tests might be inaccurate. With millions of whole-genome sequences, biologists believe, they can begin to work out those crucial combined effects.
One upcoming study shows how important gene combinations can be. In research scheduled for publication in the journal Human Molecular Genetics, scientists led by Charis Eng of the Cleveland Clinic examined the incidence of breast, thyroid, and other cancers in patients carrying a mutation in a gene called PTEN. Such mutations are typically interpreted as raising the risk of cancer. But Eng found that the presence or absence of mutations in another gene, called SDHx, can alter that risk.